Envisioned task
The aim of this track is to assess the performance of shape retrieval algorithms (deterministic and/or machine learning (ML) methods) on a large dataset of protein surfaces through inclusion of homologous structures, alternate conformations and physicochemical properties. Participants are asked to correctly assign classes to proteins.
Proteins are biological macromolecules which are classified according to their evolutionary relationship and/or their structure. They can be described as non-rigid surfaces which represent the solvent-excluded surface (Connoly et al., J Appl Cryst. 1983). They can share local or global similarities with other proteins and may be close or distant homologs. Detecting similarities and dissimilarities is important in molecular biology and in drug discovery.
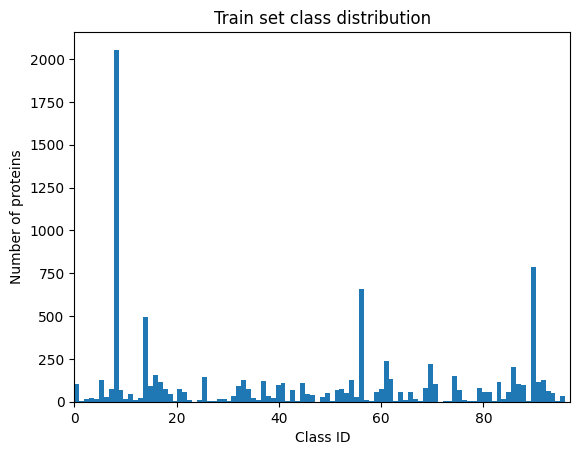
Compared to the previous SHREC Protein Shape Retrieval tracks (e.g. SHREC 2019, SHREC 2021) this track proposes a larger set of 11,565 surfaces divided into 97 imbalanced classes to simulate the reality of the biological data.
Ground truth - Training and test datasets
The ground truth is built upon the class assignment of the dataset protein surfaces based on MMSeqs2 (Steinegger et al., Nature Biotechnology 2017) clusters from the RCSB API. Only experimental models (from X-ray and NMR) with more than 50 residues have been conserved. Their solvent-excluded surfaces (SES) with physicochemical properties such as electrostatic potential were calculated.
This dataset of 11,555 surfaces belonging to 97 classes will be split into a training set and a test set, in an 80/20 proportion, respectively. The training set will contain 9,244 protein surfaces with their corresponding ground truth class, whereas the test set will be composed of 2,311 protein surfaces without annotation.

The potential is calculated with the Treecode-Accelerated Boundary Integral Poisson-Boltzmann solver using APBS (Geng, Krasny, 2013). The surface is generated with NanoShaper.
The VTK output format contains the following information: POINTS, POLYGONS, POTENTIALS and NORMAL POTENTIALS.
Update: the dataset have been updated. See the Git repository for more information.
To download the training and the test sets, and to access the raw results of the paper:
- Training_Set: Downloads VTK files — Download CSV file (paper version) — Download CSV file (updated: 28/10/25)
- Test_Set: Download VTK files — Download CSV file (paper version) — Download CSV file (updated 28/10/25)
- Ground_Truth_of_Test_Set: Download CSV file (paper version) — Download CSV file (updated: 28/10/25)
- Methods and Results: Git (methods & raw results)
- Paper published in Computers & Graphics: Paper
Evaluation
For all methods, participants are asked to produce a CSV file containing for each object:
- the ID of the object,
- the ID of the predicted class.
Be careful that the IDs of the predicted classes correspond to the IDs given in the ground truth training set. If some methods produce also a dissimilarity matrix before class assignment, they may also provide this matrix. The ground truth (class annotation) will be used to evaluate the class prediction.
Participants are also asked to provide runtimes and hardware for their calculations.
The performance will be assessed by using classification measures and statistics.
Schedule Timeline & Registration
- March 14, 2025: Dataset is made available, and participants are allowed to run their calculations.
- April 4, 2025: Registration deadline. Registration must be sent to Taher Yacoub, Camille Depenveiller and Matthieu Montès.
Register here - April 14, 2025: Submission deadline of the results to the organizers. Participants are also asked to send a summary of the method and results to be included in the track report. A link to download/compile the method is expected as well.
- April 15, 2025: Ground truth release to the participants.
- April 30, 2025: Submission deadline of the paper.
- September 4-5, 2025: Eurographics 2025 Symposium on 3D Object Retrieval.
Expected participants
All 3DOR experts who are interested in treating non-conventional shapes with inherent complexity and who want to evaluate ML and/or deterministic methods.
Register hereOrganizers
For any problems or information, do not hesitate to contact us.Taher Yacoub - Conservatoire National des Arts et Métiers - taher.yacoub@lecnam.net
Camille Depenveiller - Conservatoire National des Arts et Métiers - camille.depenveiller@lecnam.net
Matthieu Montès - Conservatoire National des Arts et Métiers - matthieu.montes@cnam.fr
